Raymond - ML MRI Tumor Classifier
 Raymond Damadian
Raymond DamadianThis project identifies and classifies tumors in the brain based on images taken using an MRI machine. I have trained 17 machine learning models over 2 datasets consisting of MRI brain scans.
Jump to Source CodeWho is Raymond Damadian?

Raymond Damadian is an American physician, scientist, and inventor who is best known as the father of the MRI Machine.
Damadian's groundbreaking work in the field of medical imaging led to the development of the MRI, which revolutionized the way doctors diagnose and study diseases.
Apart from his work on MRI technology, Damadian has made contributions in other areas of medicine, including his research on the role of MRI in detecting and diagnosing cancer.
Raymond Damadian's work has had a profound impact on the field of medicine and has helped improve the diagnosis and treatment of numerous medical conditions. His pioneering efforts in the development of MRI technology have saved countless lives and continue to be a vital tool in modern healthcare.

Raymond Damadian | Biography
Raymond Damadian was the first scientist to present data regarding radio frequency signals in tissues.
What is this project about?
This project identifies and classifies tumors in the brain based on images taken using an MRI machine. I have trained 17 machine learning models, over 2 datasets consisting of MRI brain scans. 15 models on dataset 1 consisting of 5 types of image filtering (canny, harris, kMeans, hough, no change) across 3 imaging modalities (t1, flair, segmentation). 2 models on dataset 2 consisting of 2 types of image filtering (kMeans, no change)
My key finding was that filtering doesn’t help with machine learning, since filtering will help a human determine important information by removing noise. While for a machine, it is not the best since I’m just limiting the amount of diverse data it can learn on. Canny and Harris change the image to black and white which reduces the image’s “diversification”. Though, this could be because my ML is still very early stage. N.b I haven’t done any machine learning before this, so lots of my data is potentially over or underfit.
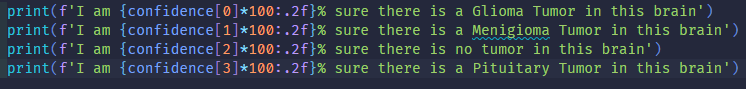
Set 1 is a data set used for binary classification, whether the brain does or doesn’t contains an abnormal mass of tissue, while set 2 is another dataset used for multi-class classification, whether the brain has a glioma, meningioma, pituitary tumor or no tumor.
All the models are located in the Github repository, the model used in the live website is the model trained on dataset 2 with no filtering, since it performed the best.
I chose this as a way to tackle the important topic of cancer in the brain. To educate me on this important topic and to explore Convolutional Neural Networks. The key challenge was taking in a lot of information, I went over the Google Crash Course on Machine learning and still felt like I didn’t have enough knowledge to tackle this topic. I had so many challenges, here’s a list:
- Collecting enough data, finding a dataset that addressed modalities
- Preventing Overfitting and co-adaptation neurons via Dropout Layers
- Finding a good way to evaluate the model when placed against my test set
- Modeling the model’s performance and identifying whether or not it was under or overfit
- Knowing what layers to use to build my CNN model (Dense Layers, Convolution Layers, Pooling Layers)
- Knowing what typing of activation functions to use for each artificial neuron (Sigmoid, Relu, Leaky Relu, etc.)
- How to split my data set into training, validation, and testing
- Choosing a batch size for each iteration
These were challenges and still are challenges since I still don’t have a full understanding of them and just referred to material online and looked at what many other people have done.
Source Code
MRI Brain Tumor Classifier | Github Repository
Project used to explore different kind of method of machine learning and computer vision to detect brain tumor in MRI scans.
Background Information
I didn’t do any research on papers, though I did look at someone’s prior work on Kaggle who trained a generalized CNN with 96% Accuracy on set2 and even works very well on generalized data while my models don’t allow for broad/diverse input, which isn’t great.
This Notebook by MD Mushfirat Mohaimin was very thorough, he/she performed data scaling (which I also did) for decreased training time, dataset balance analysis, data augmentation (varying the brightness and contrast of the data), and trained the model on the VGG16 CNN architecture.
But I looked at his/her work after I finished most of the project. So I didn’t consider any preexisting architectures and just fixed my layers to 3 Convolutional layers (16, 32, 16 neurons) 3 Pooling Layers to Downsample, a flattening layer, and finally 2 dense layers (256 neurons outputting to either 4 or 1 neuron depending on binary or categorical classification)
Dataset Information
Set1 consisted of 3540 png images split into 2 classes (per modality)
Set2 consisted of 7023 jpeg images split into 4 classes
First I split each image into its proper classes, the images were labeled but not placed into the right folder, so I used the python os library to do some organization. Next, I load the data (shuffled) using the Keras Util library and scaled the data so each pixel value is between 0 and 1, so it’s easier and faster to train the model. Then, split the data into 3 sets, the training set (70%), the validation set (20%), and the test set (10%).
After, we built our model as stated below:
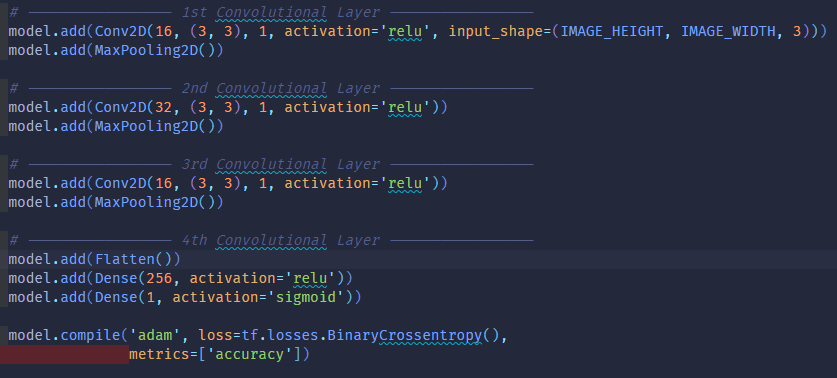
Once the model is built, we pass in our training data and validation data to train/fit the model to the data. I used the early stopping callback to prevent overfitting (validation vs training divergence) by monitoring the decrease of validation loss with a patience value of 5 increase epochs, where validation loss is increasing. Finally, I evaluate the model on my testing data; looking at 3 metrics; precision, recall, and binary accuracy to figure out how my model might perform against real images.
Results of My Training

Project Link

Raymond | MRI Brain Tumor Classifier
MRI Brain Tumor Classifier - Project used to explore different kind of method of machine learning and computer vision to detect brain tumor in MRI scans.
Retrospective
Based on the findings, my models can’t be used in a real-life setting since my validation curves and training curves diverge, they all have a pretty good chance of being under or overfit. Furthermore, I feel like the segmentation modality, isn’t a true modality and the scan has been segmented by a professional which is why they have trained so well, thus models trained on the segmentation dataset might be exposed to label leakage. If the segmentation type is an actual modality with no human manipulation, then you might be able to use this in the real world since it performs so well, maybe…
This was more of an experiment to test whether image filters such as canny, harris, hough circle, and k-means can aid CNNs in learning the right features of the image. At least in this experiment, I can conclude that it didn’t help based on the way they were filtered. K-means Clustering for color quantization seems like it helped the most out of all the other types of filtering since it yields similar image results to the segmentation modality. In terms of future work, it would be great to try more types of filtering, changing the parameters of the filters used. And experimenting with more Convolutional neural network architectures (Such as PolyNet, VGG, Xception, and ZfNet) and hyperparameters within my current model.
Appendix
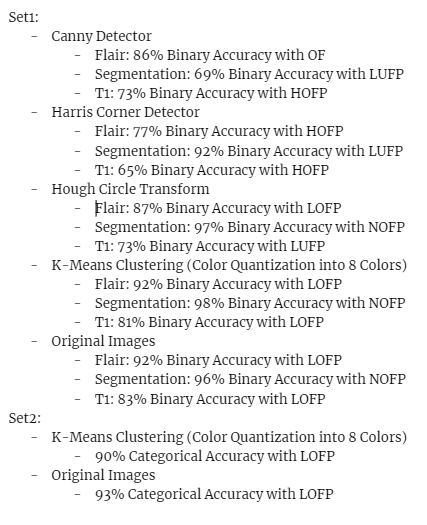
1.1 Binary Classification Results
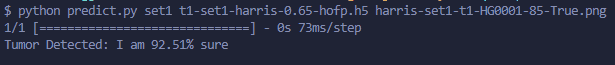

1.2 Binary Returned Logic

1.3 Multi-Class Classification Results
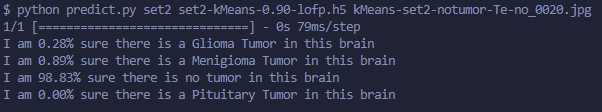
1.4 Multi-Class Returned Logic
What is the meaning behind the project name?
I've written a post about the meaning behind my project names, you can read about it here.
Built with Next.js and Tailwind CSS. Made with ❤️ by Justin Zhang.